Abstract
The success of modern, evidence based and personalized medical research is highly dependent on the availability of a sufficient data basis in terms of quantity and quality. This often also implies topics like exchange and consolidation of data. In the area of conflict between data privacy, institutional structures and research interests, several technical, organizational and legal challenges emerge. Coping with these challenges is one of the main tasks of information management in medical research. Using the example of cancer research, this case study points out the marginal conditions, requirements and peculiarities of handling research data in the context of medical research.
Introduction
First the general importance of data exchange and consolidation will be discussed. In the second section, the important role of the patient in medical research will be addressed and how it affects the handling of data. The third section focuses on the question what the role of open data could be in this context. Finally, the fourth section tackles the topic of challenges of open data in the context of medical (research) data. It tries to illustrate why it is a problem and what the obstacles are.
Importance of Data Exchange and Consolidation
With oncology striving after personalized medicine and individualized therapy, stratification becomes a major topic in cancer research. The stratification of tumor biology and patients is important to provide individual therapies with maximum tumor control (optimally total remission) and minimal side effects and risks for the patient. Therefore, the search for diagnostic markers, e.g. diagnostic imaging, antibody tests or genome analysis, as well as for adequate treatments with respect to specific markers is constantly intensified.
Looking at research results, it becomes obvious that cancer diseases (e.g. prostate cancer or breast cancer) are more like disease families with a multitude of sub-types and that the anatomical classification of tumors might be misleading and a classification according to the pathological change of signaling pathways on the cellular level is more adequate. This differentiation is very relevant because for one patient a certain treatment may be effective and absolutely relevant while it has no positive impact on tumor control for other patients with the “same” cancer and only bears side effects.
In order to have an evidence-based medicine with a sound statistical basis, the amount and quality of available data becomes very important. The required amount of data increases with the number of relevant factors. Looking at the current cancer research, one has a vast array of factors and information—and it is still increasing. One has for example the patient and tumor biology (e.g. a multitude of diagnostic images; analysis of the genome, proteome etc; lab results; cell pathologies; …); way of living before and after the diagnose/therapy; environmental factors and chosen individual therapy.
The current situation can therefore be characterized as too few cases for too many factors. The size of sample sets even large institutions can collect is too small for evidence-based and highly stratified medicine. John Wilbanks, the chief commons officer of Sage Bionetworks1, put this fact more bluntly:
[…] neither Google nor Facebook would make a change to an advertising algorithm with a sample set as small as that used in a Phase III clinical trial. John Wilbanks, Sage Bionetworks. Kotz, J.; SciBX 5(25); 2012
One strategy to tackle these shortcomings is to build up networks and initiatives and pool the data to acquire sufficient sample sets2. This is not a trivial task because of the heterogeneity of the public health sector that has to be managed. You have got several stakeholders, heterogeneous documentation of information (different in style, recorded data, formats, storage media) and different operational procedures (time and context of data acquisition).
Thus, it is inevitable to cope with this heterogeneity and to build large study bases by sharing and pooling medical research data in order to realize evidence-based personalized medicine. One way to achieve this goal could be the adaption of ideas and concepts of open research data (see below).
Role of the Patient and Its Data
As described in the previous section, data is of high importance. This data cannot be collected without patients and their cooperation is crucial on several levels. This leads to a very central role for the patient and, in addition, to a special nature of medical data and its acquisition compared to other research data.
1. Medical data is personal data
By default medical data is always personal data. The implications that derive from this fact may vary according to the legal framework of a country (e.g. USA: Health Insurance Portability and Accountability Act (HIPAA); Germany: right to informational self-determination/personal rights), but it has almost always an impact on how medical data may be acquired, stored and used. In Germany, for instance, an individual (in this context a patient) must always be able to query which personal information is stored, where the information is stored and for which purpose this information is used. The information may only be altered, transferred, used, stored or deleted with according permission and sufficient traceability guaranteed.
2. Ethics
Having an experimental setup that allows the acquisition of data suitable for verifying or falsifying the scientific hypothesis goes without saying. But in the context of human research it is also mandatory to ensure that ethical principles are regarded. These principles are often derived from the Declaration of Helsinki3 and implemented by national regulations (e.g. USA: institutional review boards; Germany: Ethikkommision). Thus every study design is reviewed and needs ethic approval. This may lead to situations where experimental setups are optimal from a technocratic research perspective but cannot be approved ethically and therefore must be altered or not conducted.
3. Lack of predictability and limits of measurements
Most research-relevant incidents (e.g. (re)occurrence of an illness, adverse reactions) are not predictable and not projectable (fortunately; see “Ethics”). Therefore, you have to wait until enough natural incidents have happened and are monitored. The latter can be complicated, because not every measurement technique can be used arbitrarily frequent due to technical, ethical4 or compliance5 reasons. Without the possibilities to repeat6 “measurements” and in conjunction with the heterogeneity explained in the previous section, getting a sufficient number of cases is a nontrivial task.
4. Long “field” observation periods
In order to derive conclusions that really matter for patients, like “improved survival”or “improved quality of life” you need observation periods of 10 and more years. In this time, the individuals will move around in the distributed public health system (e.g. by changing their place of residence, choosing new general practitioners). Data will be accumulated, but is not centrally available because of the heterogeneous nature of the system. Therefore, keeping track on a study participant and assembling a non-biased, non-filtered view on study relevant data7 can be very complicated.
5. Compliance
Besides all the explained technical and organizational problems, the key stakeholder is the study participant / patient and its compliance to the study and the therapy. If the participant is not compliant to the study, he drops out, which results in missing data. This missing data can lead to a selection bias and must be handled with expertise in order to make a reliable assessment of the trial’s result. The dropout rates vary and depend on the study; rates around 20% are not unusual, also rates up to 50% have been reported.
Participants that are not therapy compliant alter the therapy or skip it totally (e.g. changing medication; skipping exercises; taking additional drugs). According to a report (WHO 2003) of the World Health Organization up to 50% of the patients are not therapy compliant. An unnoticed lack of therapy compliance may introduce a bias towards the trial results.
6. Consent
The patient has to consent8 on three levels before he can be part of a medical trial. First, he must consent to a therapy that is relevant for the trial. Second, if all inclusion criteria and no exclusion criteria for the trial are met, the patient must consent to be part of the trial. Third, the patient must consent to the usage of the data. The third consent exists in different types, namely: specific, extended, unspecific/broad. The specific consent limits the usage to the very trial it was made for. In the context of open data this type of consent is not useful and is considered as limiting by many researchers (see challenges). The extended consent often allows the usage for other questions in the same field as the original trial (e.g. usage for cancer research). If it is extended to a level where any research is allowed, it is an unspecific consent. An example for this type of consent is the Portable Legal Consent devised by the project “Consent to Research”9.
You may find each aspect in other types of research data, but the combination of all six aspects is very distinctive for medical research data and makes special handling necessary.
Role of Open Research Data
The chapter “Open Research Data: From Vision to Practice” in this book gives an overview over the benefits open data is supposed to bring. Websites like “Open access success stories”10 try to document these benefits arising from open access/open science. Also in the broad field of medical research, many groups advocate a different handling of data (often in terms of open data).
One main reason is the requirement of transparency and validation of results and methods. For example in the domain of medical image processing the research data (test data, references and clinical meta data) is often not published. This renders the independent testing and verification of published results, as well as the translation into practice very difficult. Thus initiatives like the concept of Deserno et al. (2012) try to build up open data repositories. Another example would be the article of Begley and Ellis (2012), which discusses current problems in preclinical cancer research. Amongst others, it recommends a publishing of positive and negative result data in order to achieve more transparency and reliability of research.
Besides this, several groups (e.g. the Genetic Alliance or the former mentioned project Consent to Research) see open access to data as the only sensible alternative to the ongoing privatization of science data and results. For instance the company 23andMe offers genome sequencing for 99$. In addition to the offered service the company builds up a private database for research and the customers consent that this data may be used by the company to develop intellectual property and commercialize products11.
Another topic where the research community could benefit from the implementation of open data publishing is the heterogeneity of data (see next section: challenges). Making data available means, that it is:
- open (in terms of at least one public proceeding to get access)
- normed (content of data and semantics are well defined)
- machine readable
- in standardized format
Having this quality of data would be beneficial, for instance, for radiology, whose „[…] images contain a wealth of information, such as anatomy and pathology, which is often not explicit and computationally accessible […]“, as stated by Rubin et al. (2008). Thus, implementing open data could be an opportunity to tackle this problem as well.
Challenges
The previous sections have discussed the need for data consolidation, the peculiarities of medical research data and how medical research is or could be (positively) affected by concepts of open research data. It is irrelevant which approach is taken in order to exchange and consolidate data, you will always face challenges and barriers on different levels: regulatory, organizational and technical.
The general issues and barriers are discussed in detail by Pampel and Dallmeier-Tiessen (see chapter 14, Open Research Data: From Vision to Practice). This section adds some aspects to this topic from the perspective of medical research data.
Regulatory constraints for medical (research) data derive from the necessity of ethic approval and legal compliance when handling personal data (see section 2, point 1 and 2). There are still open discussions and work for the legislative bodies to provide an adequate frame. The article of Erika C. Hayden (2012a) depicts the informed consent as a broken contract and illustrates how today on one hand participants feel confused by the need of “reading between the lines”, on the other hand researchers cannot pool data due to specific consents and regulatory issues.
Although there are open issues on the regulatory level, ultimately it will be the obstacles on the organizational and technical level – which may derive from regulatory decisions – which determine if and how open data may improve medical research. Therefore, two of these issues will be discussed in more detail.
Pooling the Data
Given that the requirements are met and you are allowed to pool the data of different sources for your medical research, you have to deal with two obstacles: mapping the patient and data heterogeneity.
As previously noted, patients move within the public health system and therefore medical records are created in various locations. In order to pool the data correctly, you must ensure that all records originated with an individual are mapped towards it but no other records. Errors in this pooling process lead either to “patients” consisting of data from several individuals or the splitting of one individual in several “patients”. Preventing these errors from happening can be hard to implement because prevention strategies are somehow competing (e.g. if you have very strict mapping criteria, you minimize the occurrence of multi-individual-patients but have a higher change of split individuals due to typing errors in the patient name).
In the case that you have successfully pooled the data and handled the mapping of patients, the issue of heterogeneity remains. This difference of data coverage, structure and semantics between institutions (which data they store, how the data is stored and interpreted) makes it difficult to guarantee comparability of pooled data and to avoid any kind of selection bias (e.g.: Is an event really absent or just not classified appropriately by a pooled study protocol).
Anonymization, Pseudonymization and Reidentification
Individuals must be protected from (re)identification via their personal data used for research. German privacy laws, for instance, define anonymization and pseudonymization as sufficient, if they prohibit reidentification or reidentification is only possible with a disproportional large expenditure of time, money and workforce12. Ensuring this requirement becomes increasingly harder due to technical progress, growing computational power and – ironically – more open data.
Reidentification can be done via data-mining of accessible data and so-called quasi-identifiers, a set of (common) properties that are – in their combination – so specific that they can be used to identify. A modern everyday life example would be Panopticlick. It is a website of the Electronic Frontier Foundation that demonstrates the uniqueness of a browser (Eckersley 2010) which serves as a quasi-identifier. Therefore, a set of “harmless” properties is used, like screen resolution, time zone or installed system fonts.
The following examples illustrate possibilities and incidents of reidentification:
- Simple demographics. The publications of Sweeney (2000) and Golle (2006) indicate that for 63%–87% of the U.S. citizens the set of birth date, sex and postal code is unique and a quasi-identifier.
- ICD codes. Loukides et al. (2010) assume that 96.5% of the patients can be identified by their set of ICD913 diagnoses codes. For their research the Vanderbilt Native Electrical Conduction (VNEC) dataset was used. The data set was compiled and published for an NIH14 funded genome-wide association study.
- AOL search data. AOL put anonymized Internet search data (including health-related searches) on its web site. New York Times reporters (Barbaro et al. 2006) were able to re-identify an individual from her search records within a few days.
- Chicago homicide database. Students (Ochoa et al. 2001) were able to re-identify a 35% of individuals in the Chicago homicide database by linking it with the social security death index.
- Netflix movie recommendations.15 Individuals in an anonymized publicly available database of customer movie recommendations from Netflix are re-identified by linking their ratings with ratings in a publicly available Internet movie rating web site.
- Re-identification of the medical record of the governor of Massachusetts. Data from the Group Insurance Commission, which purchases health insurance for state employees, was matched against the voter list for Cambridge, re-identifying the governor’s health insurance records (Sweeney 2002).
The examples illustrate the increasing risk of reidentifiaction and the boundary is constantly pushed further. If you look for example at the development of minaturised DNA sequenzing systems16 (planned costs of 1000 US$ per device), sequencing DNA (and using it as data) will presumably not stay limited to institutions and organisations who can afford currently expensive sequencing technologies.
Thus proceedings that are compliant to current privacy laws and the common understanding of privacy are only feasible if data is dropped or generalized (e.g. age bands instead of birth date or only the first two digits of postal codes). This could be done for example by not granting direct access to the research data but offering a view tailored for the specific research aims. Each view ponders the necessity and usefulness of each data element (or possible generalizations) against the risk of reidentification.
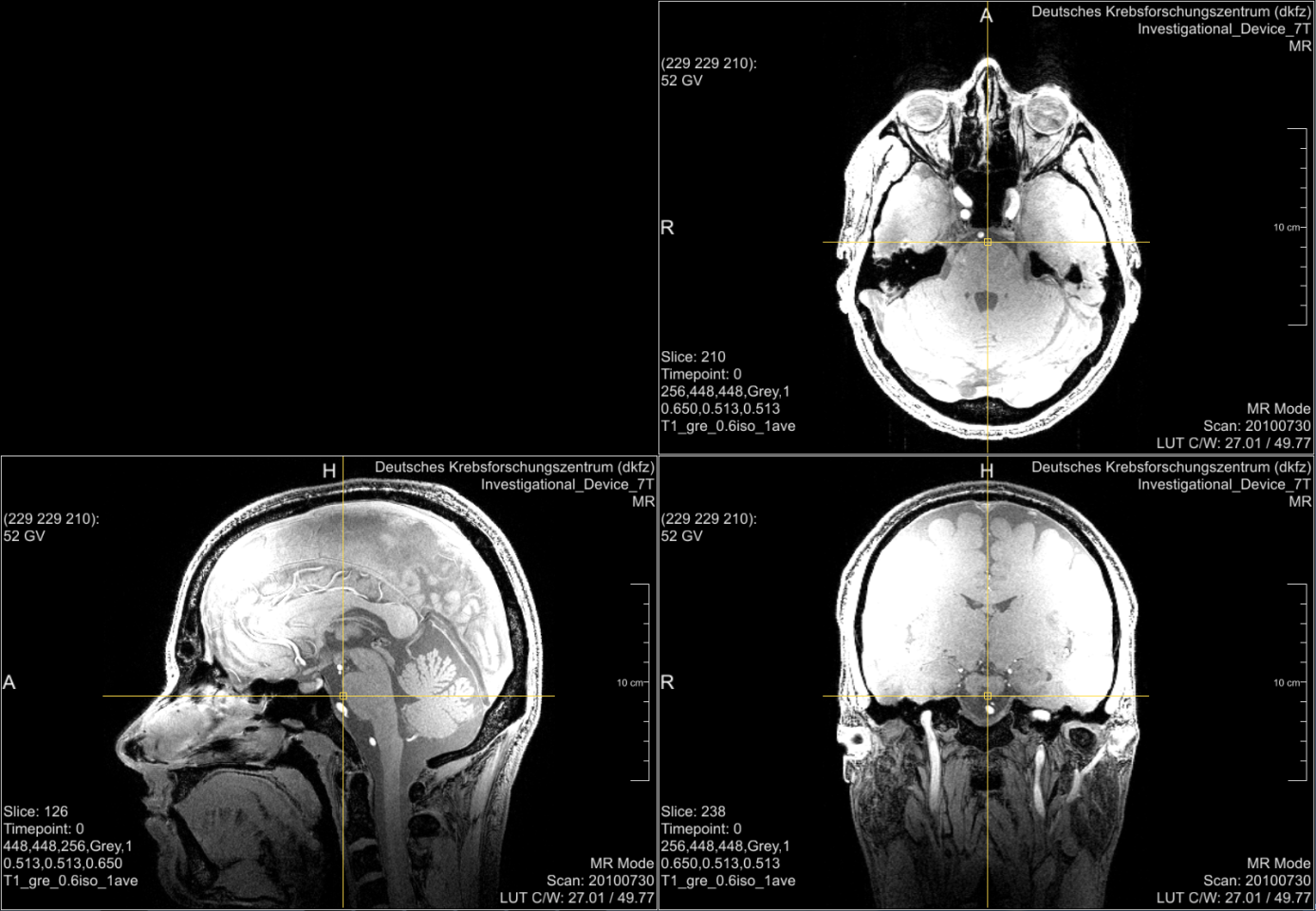
Even if an infrastructure is provided that enables the filtering of data described above, you will always have medical data that is easily reidentifiable and at least hard to be pseudonymized. Good examples are radiological head or whole body images. Figure 1 shows head images from a study participant17. The original perspective of the image (axial view) and the other medical perspectives (sagittal and coronal view) may not be suitable for reidentification by everyman. But a simple volume rendering of the data (Figure 2) allows easy reidentification. Starting from this point with modern technologies several scenarios are not too far-fetched. An artificial picture, for instance, could be reconstructed and used with available face recognition APIs18 or you could take the volume data convert it into a 3D model and print it via a 3D-printer19.

References
Barbaro, M. et al., 2006. A face is exposed for AOL searcher no. 4417749. NY Times.
Begley, C.G. & Ellis, L.M., 2012. Drug development: Raise standards for preclinical cancer research. Nature, 483(7391), pp.531–533. doi:10.1038/483531a.
Check Hayden, E., 2012a. Informed consent: A broken contract. Nature, 486(7403), pp.312–314. doi:10.1038/486312a.
Check Hayden, E., 2012b. Nanopore genome sequencer makes its debut. Nature. doi:10.1038/nature.2012.10051.
Deserno, T.M., Welter, P. & Horsch, A., 2012. Towards a Repository for Standardized Medical Image and Signal Case Data Annotated with Ground Truth. Journal of Digital Imaging, 25(2), pp.213–226. doi:10.1007/s10278-011-9428-4.
Eckersley, P., 2010. How Unique Is Your Browser? In Proceedings of the Privacy Enhancing Technologies Symposium (PETS 2010). Springer Lecture Notes in Computer Science.
Golle, P., 2006. Revisiting the uniqueness of simple demographics in the US population. In WPES 2006 Proceedings of the 5th ACM workshop on Privacy in electronic society. New York, NY, USA: ACM, pp. 77–80.
Loukides, G., Denny, J.C. & Malin, B., 2010. The disclosure of diagnosis codes can breach research participants’ privacy. J Am Med Inform Assoc, 17, pp.322–327.
Ochoa, S. et al., 2001. Reidentification of individuals in Chicago’s homicide database: A technical and legal study. In Massachusetts Institute of Technology.
Rubin, D.L. et al., 2008. iPad: Semantic Annotation and Markup of Radiological Images. In AMIA Annu Symp Proc. pp. 626–630.
Sweeney, L., 2002. k-Anonymity: A Model for Protecting Privacy. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems, 10(05), pp.557–570. doi:10.1142/S0218488502001648.
Sweeney, L., 2000. Uniqueness of Simple Demographics in the U.S. Population, LIDAPWP4. In Pittsburgh, USA: Carnegie Mellon University, Laboratory for International Data Privacy.
WHO, 2003. Report. Adherence to Long-Term Therapies: Evidence for Action, Available at: http://www.who.int/chp/knowledge/publications/adherence_report/en/.
Sage Bionetworks is the name of a research institute which promotes biotechnology by practicing and encouraging open science. It is founded with a donation of the pharmaceutical services company Quinitles. cf. http://en.wikipedia.org/wiki/Sage_Bionetworks↩
An Example is the German Consortium for Translational Cancer Research (Deutsches Konsortium für Translationale Krebsforschung, DKTK; http://www.dkfz.de/de/dktk/index.html. One objective in the DKTK is the establishement of a clinical communication platform. This platform aims amongst others to better coordinate and standardize multi centric studies.↩
The Declaration was originally adopted in June 1964 in Helsinki, Finland. The Declaration is an important document in the history of research ethics as the first significant effort of the medical community to regulate research itself, and forms the basis of most subsequent documents.↩
e.g.: you cannot repeat an x-ray based imaging arbitrarily often, due to radiation exposition; you cannot expect a person suffering from cancer to daily lie in a MRI scanner for an hour.↩
e.g.: The payload for an imaging study can easily double the duration of an examination. This may lead to more stress for the participant and decreasing compliance.↩
Single measurements can be repeated (but this implies stress and leads to decreasing compliance; or is not ethically not compliant). But the complete course of treatment cannot be repeated; if a treatment event is missed, it is missed.↩
This could be a lot of (different) data. See for example the relevant factors from section 1.↩
The necessity for an informed consent of the patient can be derived from legal (see point 1) and ethical (see point 2) requirements. It is explained in detail here to characterize the different types of consent.↩
“Consent to Research” / WeConsent.us, is an initiative by John Wilbanks / Sage Bionetwirks with the goal to create an open, massive, mine-able database of data about health and genomics. One step is the Portable Legal Consent as a broad consent for the usage of data in research. Another step is the We the People petitition lead by Wilbanks and signed by 65,000 people. February 2013 the US Government replied and announced a plan to open up taxpayer-funded research data and make it available for free.↩
http://www.oastories.org: The site is provided by the initiative knowledge-exchange.info which is supported by Denmark’s Electronic Research Library (DEFF, Denmark), the German Research Foundation (DFG, Germany), the Joint Information Systems Committee (JISC; UK) und SURF (Netherlands).↩
The article of C. Hayden (2012a) discusses the topic of commercial usage on the occasion of the first patent (a patented gen sequence) of the company 23andme.↩
see § 3 (6) Federal Data Protection Act or corresponding federal state law↩
ICD: International Classification of Diseases. It is a health care classification system that provides codes to classify diseases as well as a symptoms, abnormal findings, social circumstances and external causes for injury or disease. It is published by the World Health Organization and is used worldwide; amongst others for morbidity statistics and reimbursement systems.↩
National Institutes of Health; USA↩
See http://www.wired.com/threatlevel/2009/12/netflix-privacy-lawsuit and http://www.wired.com/science/discoveries/news/2007/03/72963↩
e.g. the MinION^TM device from Oxford Nanopore Technologies (http://www.nanoporetech.com). See also (Hayden 2012b)↩
The data shown in Figure 1 and Figure 2 are provided by courtesy of Markus Graf (German Cancer Research Center).↩
One example would be web API offered by face.com (http://en.wikipedia.org/wiki/Face.com).↩
In order to print 3D-Models you can use services like http://www.shapeways.com or http://i.materialise.com.↩
Next chapter: On the Sociology of Science 2.0
Vladimir B. Teif
